New publication about peptide translocation
published 2025-09-03
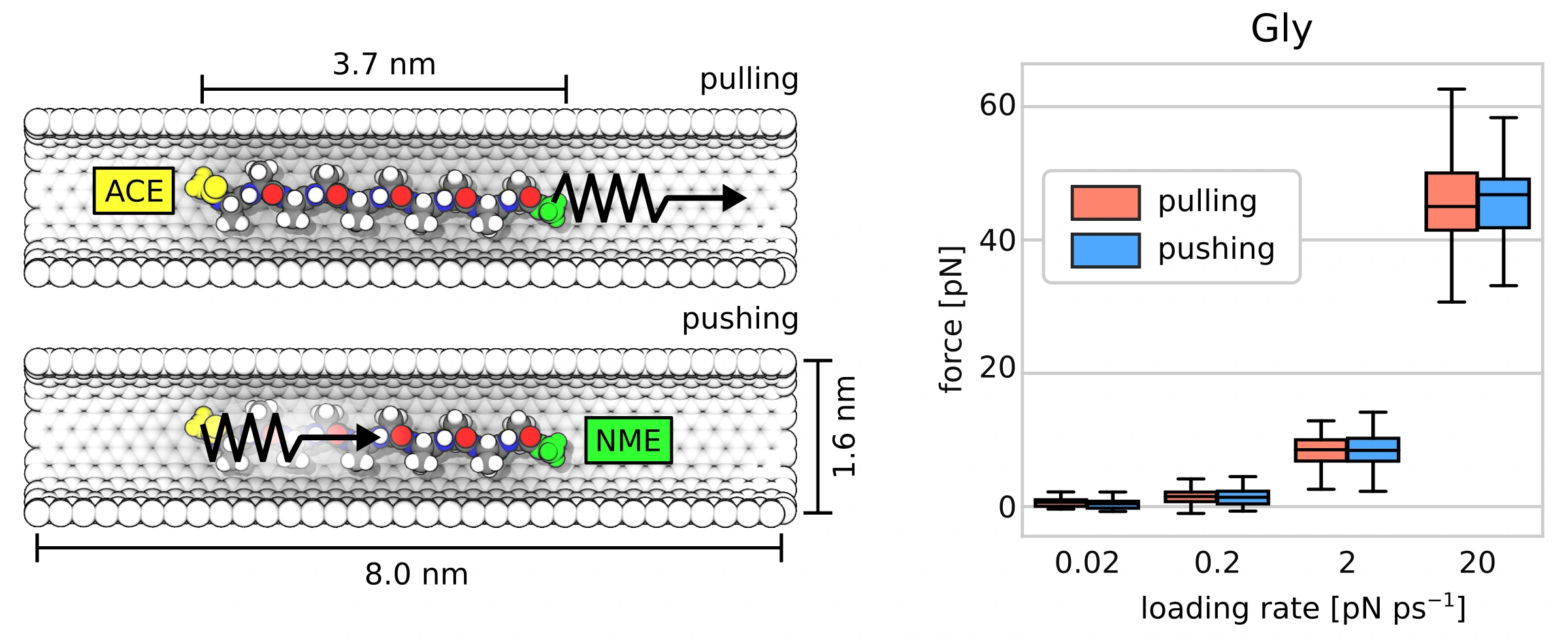
We have been studying protein synthesis on ribosomes for many years. One of the open questions concerns the translocation of the nascent protein through the ribosome during synthesis. It is still unclear what forces drive the peptide from the catalytic center, buried deep within the ribosome, toward the ribosome surface.
To address this, we performed a series of non-equilibrium molecular dynamics simulations on a simplified system. Specifically, we examined how a peptide moves through a carbon nanotube under an applied mechanical force. We were particularly interested in the difference between pulling the peptide and pushing it in the same direction. Does it matter where the force probe is attached? And would that choice affect the conformational ensemble of the peptide?
Highlights
- It's fairly easy to move a peptide through a narrow channel when an external force is applied.
- Pulling the peptide makes its movement smoother than pushing, though in both cases the peptide's own flexibility adds complexity.
- Where the force is applied matters less than the peptide's sequence or how quickly the force is applied.
full text: Physical chemistry chemical physics
preprint: https://arxiv.org/abs/2401.02352
Bachelor theses defended
published 2025-08-30
Two new chemistry graduates have successfully defended their bachelor's theses and received their degrees. Rudolf Kvasňovský and Tomáš Kuzma both gained valuable experience in setting up, running, and analyzing MD simulations in GROAMCS. Congratulations! We wish you many more successful simulations ahead.
Project retreat in Jáchymov
published 2025-07-25
Our grant from the Czech Science Foundation has been active since January. In collaboration with the Hlouchová group, we are investigating evolutionary early protein–RNA interactions. It felt like the right time to bring the two research groups together to share experimental and simulation results. We met in Jáchymov, a UNESCO World Heritage site with a long mining history. It was from here that Marie Sklodowska-Curie obtained uranium ore, leading to the discovery of the radioactive elements radium and polonium.
The two days were intense, featuring more than ten presentations, a long hike through the steep surrounding landscape, and, for most of the time, heavy rain.

Current opinion on RNA–protein interactions
published 2025-06-20
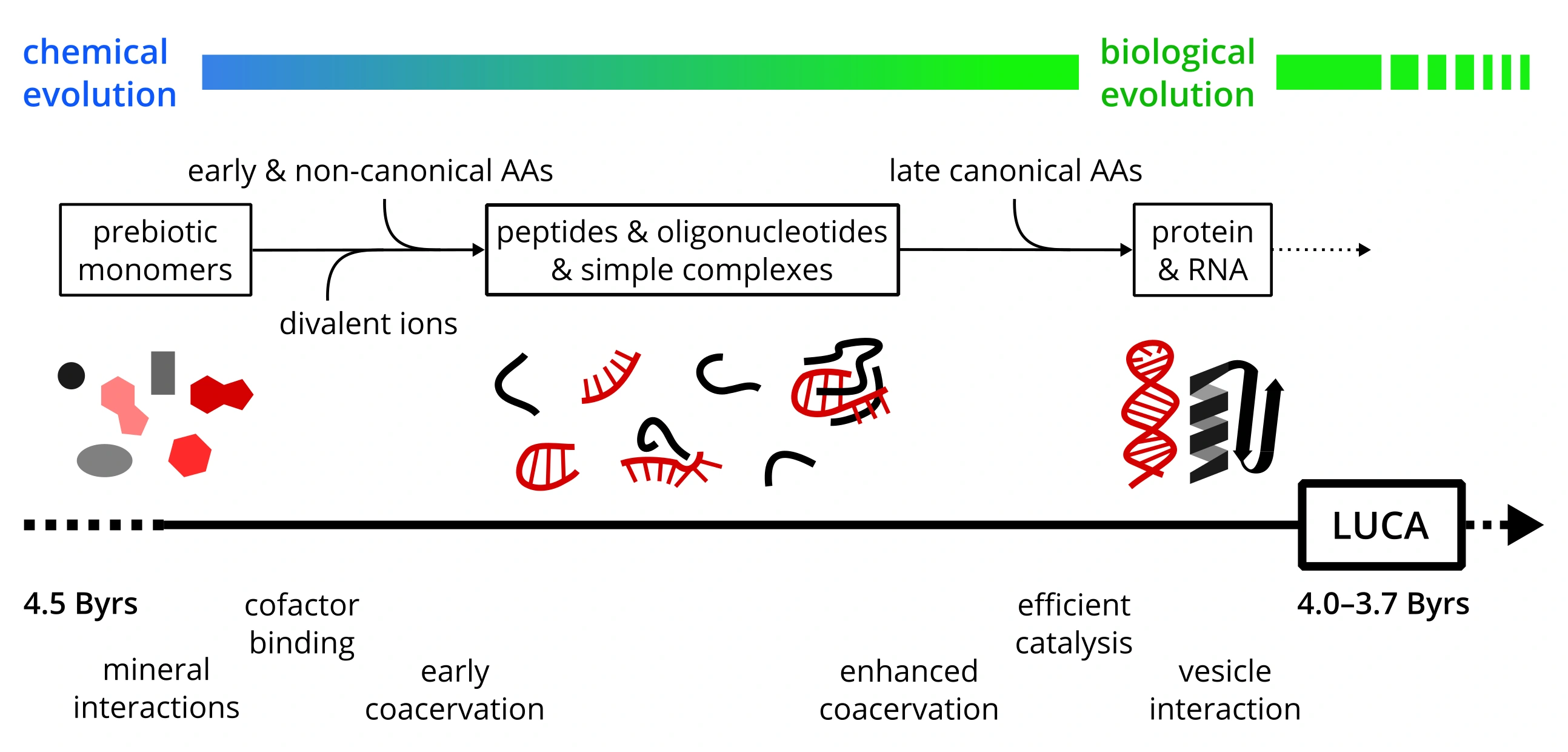
With Klára Hlouchová from Charles University in Prague, we have recently finished a review paper about the evolution of protein–RNA interactions. In fact, we have focused on the events that happened before the biological (Darwinian) evolution started to contribute.
Highlights
- The history of protein–RNA interactions reflects both prebiotic biophysical optimization and Darwinian evolution.
- The structure of extant ribosomes provides insights into the evolutionary history of protein–RNA interactions.
- Early peptide–RNA complexes could involve non-canonical amino acid residues.
- Divalent metal ions and basic amino acids played a crucial role in the emergence of protocellular assemblies.
full text: Current opinion in structural biology
preprint: https://arxiv.org/abs/2505.02037
Welcome new interns
published 2025-06-14
We are excited to welcome two new students who have joined our group for their internships.
Simone Codispoti, from the University of Milan, Italy, will be learning how to simulate protein–RNA mixtures using computational methods. As an experimental biophysicist, he will explore the molecular features of liquid–liquid phase separation in samples he previously studied during his Master's and PhD work.
We also warmly welcome Tatiana Šimeková, a talented high school student from Slovakia, who will focus on investigating unusual nascent peptides in the ribosome exit tunnel.
We’re thrilled to host you here in Prague and look forward to a productive and inspiring collaboration!

Two new engineers have graduated
published 2025-06-10
We are proud to announce that two members of our group have successfully graduated and obtained their engineering degrees from UCT Prague.
Martin Mašek completed his studies in Physical Chemistry and defended his thesis on molecular dynamics simulations of the protoribosome. He is now heading to Berlin, Germany, to begin a PhD program in computational chemistry.
Jakub Žváček graduated from the Data Engineering program, defending his thesis on the structure and dynamics of tRNA analogues. He has already joined a consulting and technology company as a data analyst.
We thank both of them for their contributions over the years and wish them all the best in their future careers.
New preprint about coarse-grained ribosome
published 2025-04-12
Our group focuses on computer simulations of ribosomes and other biomolecular complexes. We specialize in atomistic molecular dynamics simulations, where the motion of each individual atom is tracked over time. These simulation systems typically comprise around 2 million atoms, making them computationally demanding. In some cases, atomic-level resolution is unnecessary, and simpler models can be used. In such approaches, groups of atoms representing biomolecular building blocks, such as sugar moieties or nucleobases, are combined into a single coarse-grained unit called bead.
We have recently posted a preprint on simulating the bacterial ribosome using the general coarse-grained model MARTINI. Our work details the simulation protocol and highlights both the strengths and limitations of the model.
Michal has joined Czech Science Foundation
published 2025-04-03
Michal has taken on a new role at the Czech Science Foundation. Until now, he had only been a grant applicant, but starting this year, he will also be involved in evaluating grant proposals. As a member of Evaluation Panel 205 – Biophysics, Macromolecular Physics, and Quantum Optics – he will be responsible for assessing applications for scientific research funding.
The Czech Science Foundation (GACR) is an independent public organisation supporting basic research throughout all scientific fields from public funds. GACR is the only institution in the Czech Republic, which provides public funding exclusively for basic research projects.
Rudolf succeeded with his poster in Hünfeld
published 2025-03-20
Rudolf Kvasňovský, a third-year student of the bachelor's program Chemistry, won second place in the competition for the best poster at the Hünfeld 2025: Workshop on Computer Simulation and Theory of Macromolecules conference. Congratulations!
R. Kvasňovský focuses on simulations of the mitochondrial ribosome. Unlike universal ribosomes, which are found in the cytosol or on the endoplasmic reticulum of eukaryotic cells, the mitochondrial ribosome is highly specialized. In human cells, it synthesizes only 13 proteins, whereas universal ribosomes synthesize several tens of thousands of proteins. R. Kvasňovský's poster, titled Simulation Model of the Mitochondrial Ribosome, represents nearly a year of work.
Michal has joined the Editorial board of Communications Biology
published 2024-12-15
Michal will strengthen the Biophysics and Structural Biology section of the editorial board. Although he works at the Department of physical chemistry, UCT Prague, his research focuses on biomolecules and their conformational dynamics. His team investigates the conditions under which proteins are formed in cells.
Communications Biology is an Open Access journal that publishes fundamental research findings across all subfields of biology, with a strong emphasis on rigorous and rapid peer review, including options such as double-blind or open review. The editorial processing of each manuscript is handled by a global team of in-house editors and editorial board members.